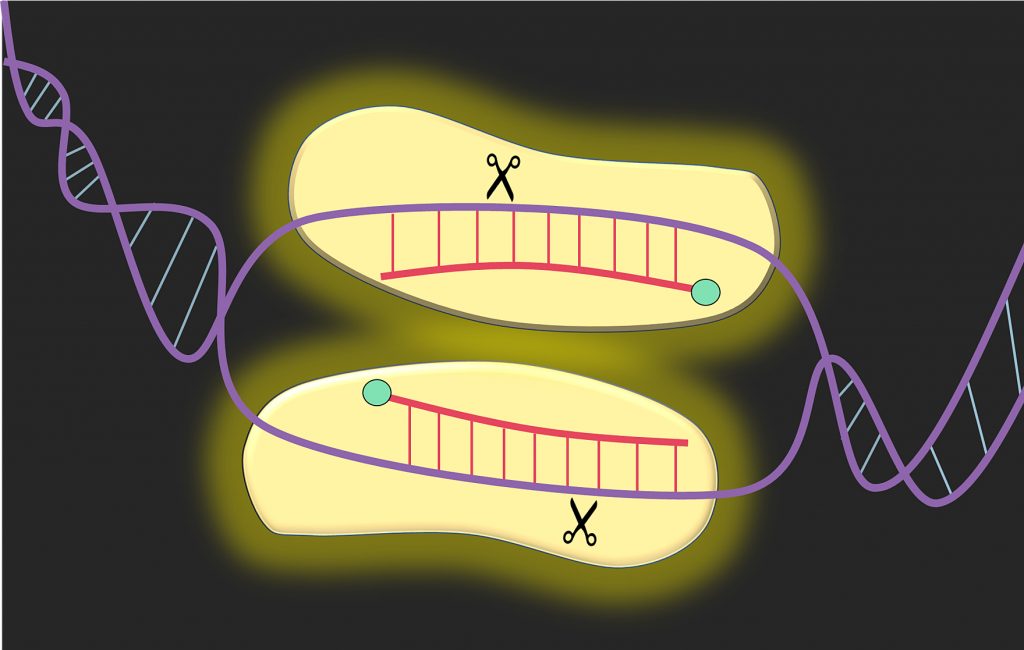
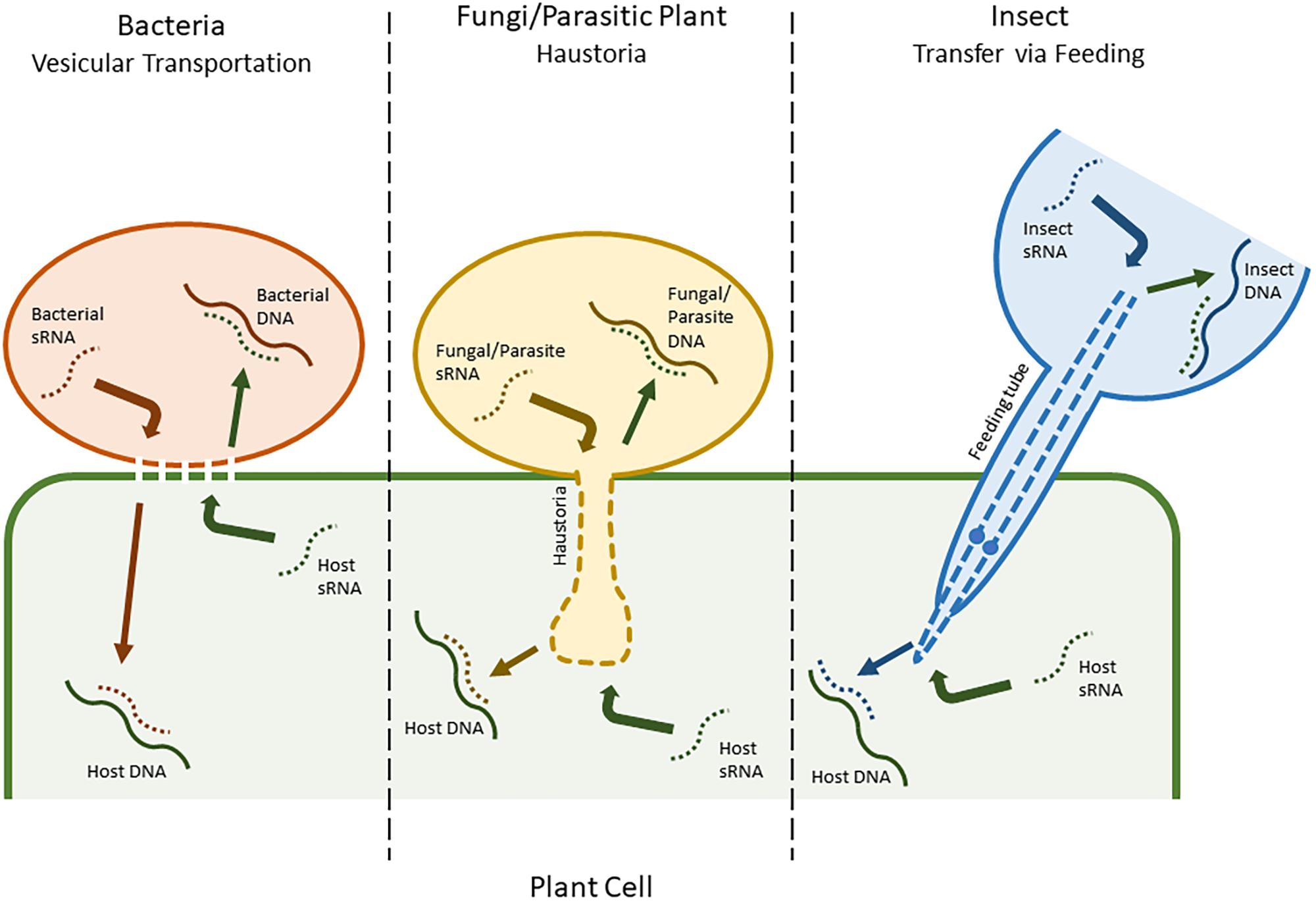
Implementing the sterile insect technique with RNA interference – a review - Darrington - 2017 - Entomologia Experimentalis et Applicata - Wiley Online Library
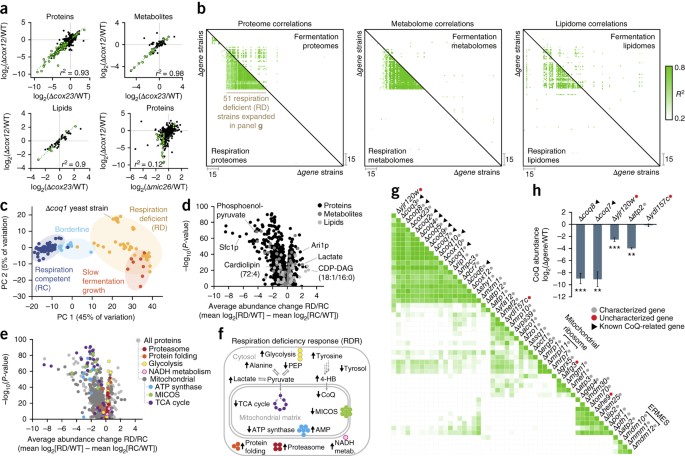
Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling | Nature Biotechnology
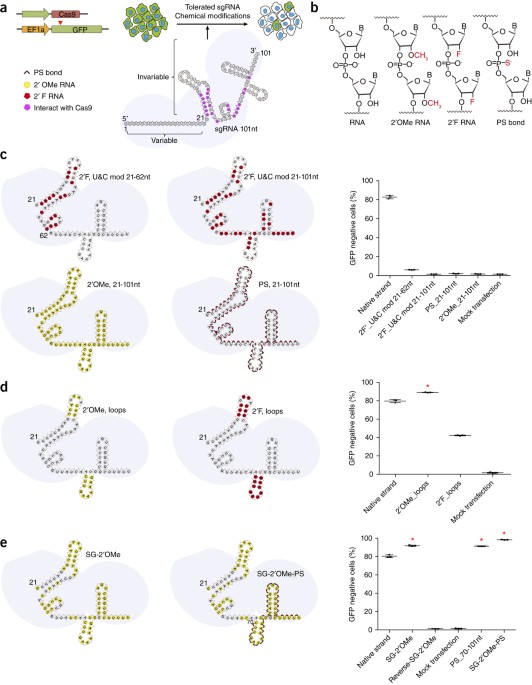
Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing | Nature Biotechnology

Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing | Nature Biotechnology

PDF) Identification of microRNAs specific for high producer CHO cell lines using steady-state cultivation

PDF) Transcript and protein expression decoupling reveals RNA binding proteins and miRNAs as potential modulators of human aging

RNA-bioinformatics: Tools, services and databases for the analysis of RNA-based regulation - ScienceDirect

Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing | Nature Biotechnology
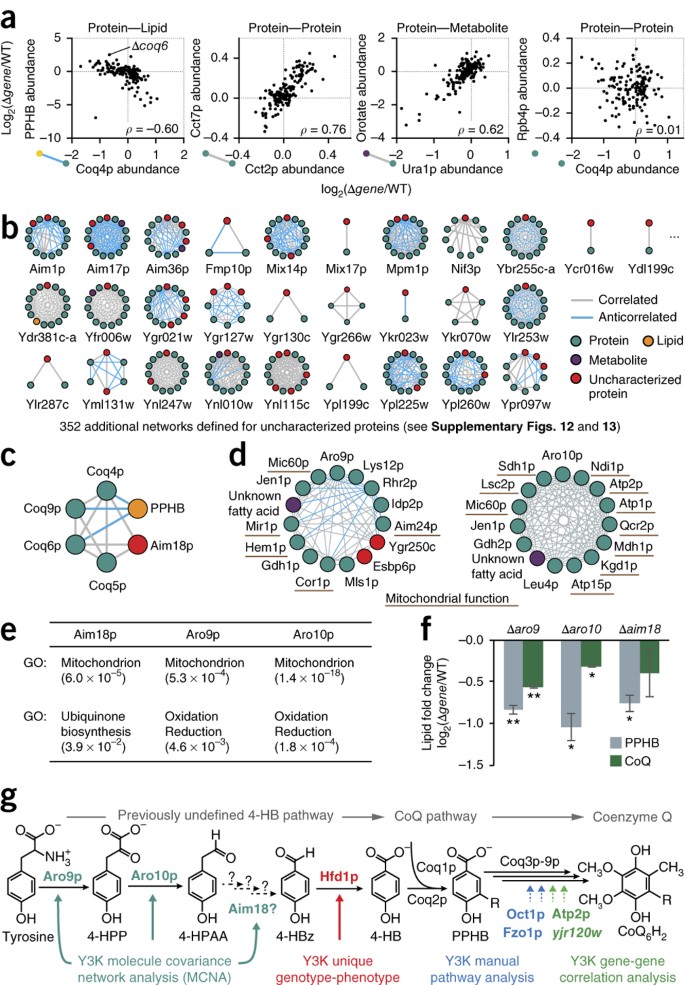
Mitochondrial protein functions elucidated by multi-omic mass spectrometry profiling | Nature Biotechnology

Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing | Nature Biotechnology
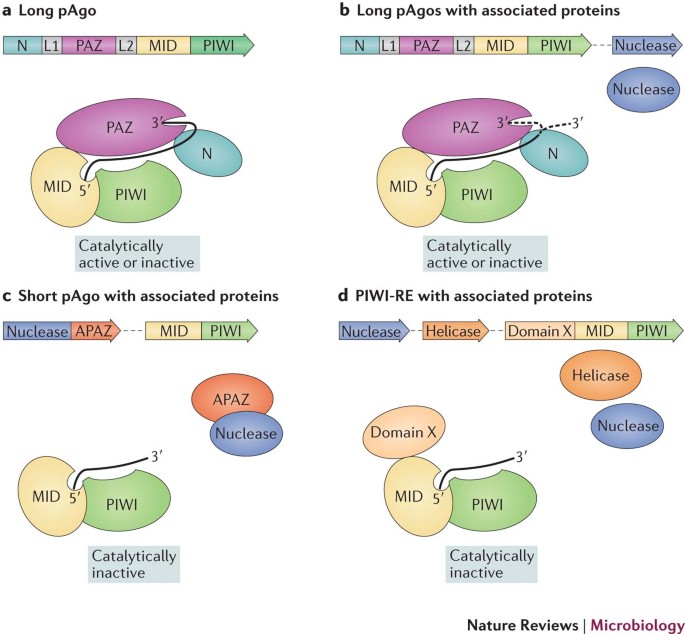
PDF) Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter

PDF) Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing

Avid Bioservices and Argonaut Manufacturing Services Enter Agreement to Offer Biotechnology and Pharmaceutical Clients Integrated Process Development, Drug Substance Manufacturing and Drug Product Parenteral Manufacturing - Argonaut Manufacturing

PDF) Anderson P, Kedersha N.. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430-436